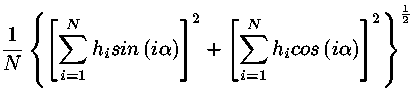
|
KEYWORD
(SHORT) |
KEYWORD
(LONG) |
DESCRIPTION |
| OFF | OFF |
Return to the main drawing mode (draw atomic coordinates).
Instead using this command, you can hit the ESCAPE key. |
| HYD | HYDROPHOBICITY | Draw averaged hydrophobicity. |
| MOM | MOMENT | Draw hydrophobic moment. |
|
PARAMETER
(SHORT) |
DESCRIPTION |
| start_index | Serial index of the first residue. |
| end_index | Serial index of the last residue. |
| COMMAND | DESCRIPTION |
|
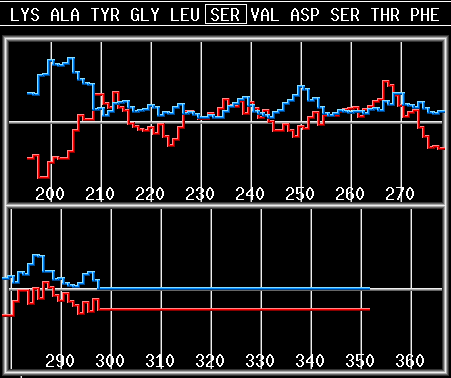
load 2por.pdb
seq from 1 scale eis win 7 angle 180 plot hyd mom 1-300 |
Load porin coordinates, copy sequence to
the main sequence buffer, select Eisenberg hydrophobicity scale, set window width to 7 and set angle to 180 degrees. Draw averaged hydrophobicity and hydrophobic moment for residues from 1 to 300. |
|
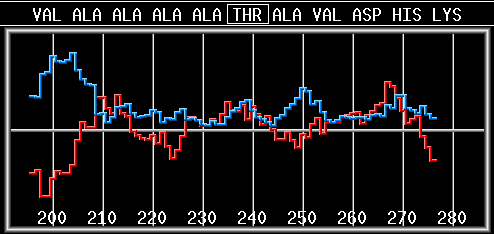
seq load sample.fasta
scale eis angle 100 plot mom |
Read the file sample.fasta, which contains
the protein sequence in FASTA format (one letter code), select Eisenberg scale, set angle to 100 and draw hydrophobic moment. |
| plot off |
Return to default drawing mode.
Instead using this command, you can hit the ESCAPE key. |