Load the first structure:
load file1.pdb
Change the color. Cyan-blue might be
a good choice:
color cyan-blue
Make the plane visible:
plane
Prepare to move the plane:
move plane
Use plane to divide structure in two parts.
The active site should be above the plane.
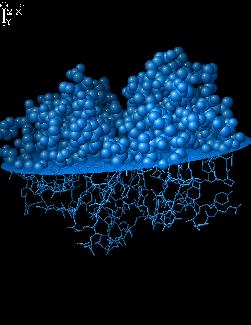
Change drawing style for atoms or bonds.
Here the spacefill style is used. This is
bad idea if using slow machine.
sel above
atom sp2
Reset movement controls:
move all
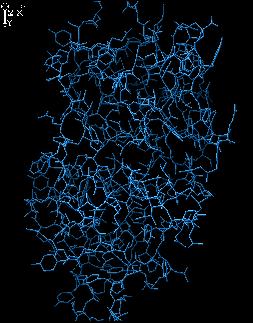
Push the first structure to the corner.
Load the second structure:
load file2.pdb
Change color. Red is recommended here:
color red
Make the plane visible and divide the
second structure in two parts. Change
drawing style for atoms or bonds.
plane
move plane
(... now rotate and translate the plane)
sel above
atom sp2
Reset movement controls:
move all
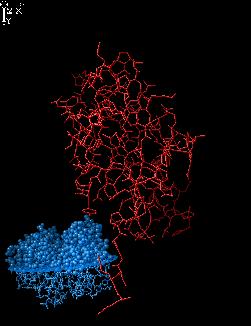
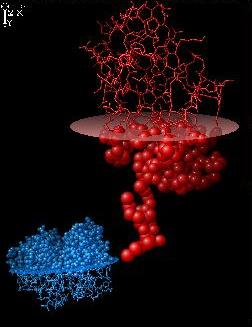
Execute DOCK command:
dock 1 2
the relative arrangement of hydrogen bond
donors and acceptors. The projections of
these side chains are given in docking
window.